My Neo4j Summer Road Trip to the World of Healthcare [Part 3]
6 min read
Part 3: Cleaning CSV Files in Bash
Hi friends and welcome back to my summer road trip through the world of healthcare. For those who are new to my adventure, this is the third part of the blog series. Catch up right here with Part 1 and Part 2.
I am using Neo4j to connect the multiple stakeholders of healthcare and hope to gain some interesting insights into the healthcare industry by the end of my exploration. This blog series demonstrates the entire process from data modeling and ETL to exploratory analysis and more. In the previous two posts, I discussed data modeling and how to integrate XML data to Neo4j by using APOC, you can find every single detail about the project on Github.
This week, I will be working with CSV files. If you are using Neo4j for the first time (like me), I can tell you honestly that loading CSV files to Neo4j is a lot easier than loading XML files. But don’t get too optimistic about it unless your data is perfectly clean. Now, let me show you the steps I used to successfully load the CSV files.
1. Get the Data
This week, our data covers information on drugs, drug manufacturers, providers and prescriptions. You can download the same data from these sources:
- FDA Drug Codes (txt, 35.1MB)
- Drug Manufacturers (txt, 1.5 MB)
- Provider Enumeration System (CSV, 5.96GB)
- Provider Prescriptions (CSV, 92.8MB)
2. Display the Data:
A. What does the data look like?
head npidata_20050523-20160612.csv
Wow, the data looks a little bit crazy, and because of that, I will not overwhelm you by copying the result here. However, I learned three characteristics about the data by displaying the first 10 rows:
- The data has a header, and the header contains white space
- The data has many columns (we will find out how many soon)
- The data has a lot of empty values
B. How many rows are in the data?
wc -l npidata_20050523-20160612.csv
Results:
4923673 npidata_20050523-20160612.csv
Each row of the data represents a registered provider in the United States from 2005 to 2016.
C. How many columns are in the data?
head -n 1 npidata_20050523-20160612.csv|awk -F',' '{print NF}'
Results:
329
Now you see my point why I said the data is a little bit crazy. But don’t panic – most of these columns do not contain values and we only need to extract a few columns to load them to my healthcare graph.
D. Remove the header from the data.
sed 1d npidata_20050523-20160612.csv > provider.csv
This will delete the first line and save the content to a new file named
provider.csv. The original file will not be changed. It’s optional to remove the header in your file because Cypher supports the ability to load a CSV file with a header and refer to the column using the header. Here is a great walkthrough tutorial of loading CSV files to Neo4j.
3. Load CSV into Neo4j
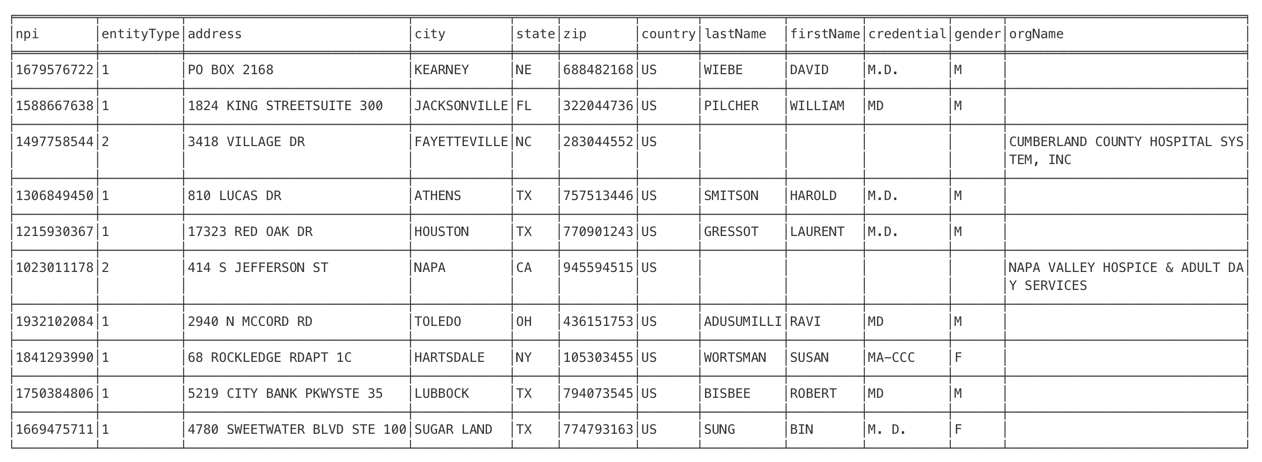
A. Display the CSV in Neo4j
LOAD CSV FROM 'file:///provider.csv' AS col RETURN col[0] as npi, col[1] as entityType, col[20]+col[21] as address, col[22] as city, col[23] as state, col[24] as zip, col[25] as country, col[5] as lastName, col[6] as firstName, col[10] as credential, col[41] as gender, col[4] as orgName limit 10
*npi: National Provider Identifier
In the figure above, I only displayed the columns that I will load into the healthcare graph.
B. Load CSV into Neo4j
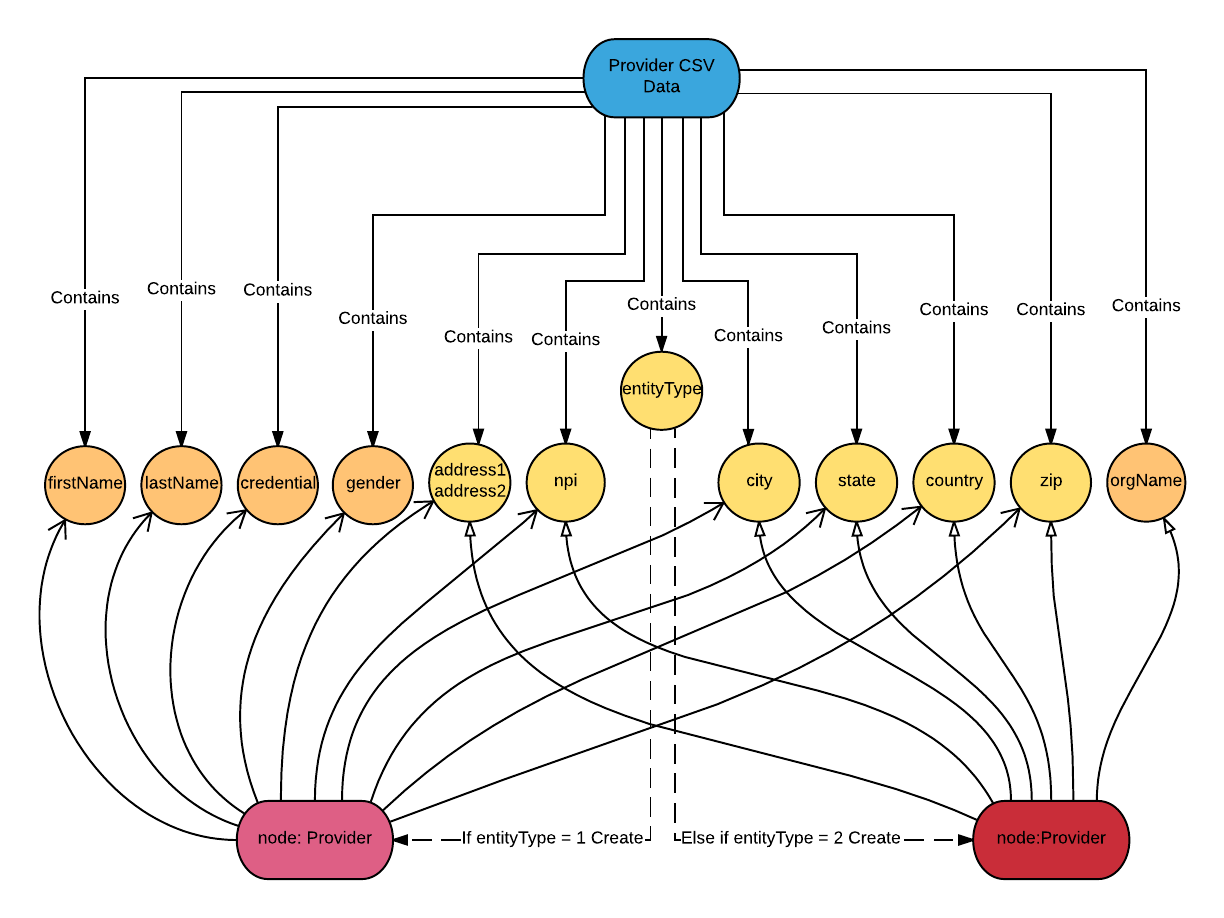
I want to create
:Provider nodes with these properties: npi, entityType, address, city, state, zip and country.When
entityType is 1, I add properties: lastName, firstName, credential and gender to the node. When entityType is 2, I add the property: OrgName to the node.Here is the Cypher query that executes the above data model and rules:
LOAD CSV FROM 'file:///provider.csv' AS col
CREATE (pd:Provider {npi: col[0], entityType: col[1], address: col[20]+col[21], city: col[22], state: col[23], zip: col[24], country: col[25]})
FOREACH (row in CASE WHEN col[1]='1' THEN [1] else [] END | SET pd.firstName=col[6], pd.lastName = col[5], pd.credential= col[10], pd.gender = col[41])
FOREACH (row in CASE WHEN col[1]='2' THEN [1] else [] END | SET pd.orgName=col[4])
The
FOREACH statement is used to mutate each element in a collection. Here I use CASE WHEN to group the data into two collections of rows: the rows with col[1] = 1 and the rows with col[1] = 2. For each row in the col[1]=1 group, I use the FOREACH statement to set the firstName, lastName, credential and gender properties, and for each row in the col[1]=2 group, I set the property orgName.C. Fix the fields containing delimiters
Running the Cypher query above returns an error:
At /Users/yaqi/Documents/Neo4j/test_0802/import/provider.csv:113696 - there's a field starting with a quote and whereas it ends that quote there seems to be characters in that field after that ending quote. That isn't supported. This is what I read: 'PRESIDENT","9'
Let’s take a look of the problematic line from the terminal:
sed -n "113697 p" provider.csv
Results:
"1790778355","2","","","BERNARD J DENNISON JR DDS PA","","","","","","","","","","","","","","","", "908 N SANDHILLS BLVD","","ABERDEEN","NC","283152547","US","9109442383","9109449334","908 N SANDHILLS BLVD"," ","ABERDEEN","NC","283152547","US","9109442383","9109449334","08/29/2005","07/08/2007", "","","","","DENNISON","BERNARD","J","PRESIDENT\","9109442383","1223G0001X","4629","NC","Y","", "","","","","","","","","","","","","","","","","","","","","","","","","","","","","","",""," ","","","","","","","","","","","","","","","","","","","","","","","","","","","","","",""," ","","","","","","","","","","","","","","","","","","","","","","","","","","","","","",""," ","","","","","","","","","","","","","","","","","","","","","","","","","","","","","",""," ","","","","","","","","","","","","","","","","","","","","","","","","","","","","","",""," ","","","","","","","","","","","","","","","","","","","","","","","","","","","","","",""," ","","","","","","","","","","","","","","","","","","","","","","","","","","","","","",""," ","","","","","","","","","","","","","","","","","","","","","","","","","","","","","",""," ","","","","","","","","","","","DR.","JR.","DDS","193400000X SINGLE SPECIALTY GROUP","","","","","","","","","","","","","",""
The problem is there is a
\ character inside the field PRESIDENT\, and when loading the file, Cypher will skip the double quotation followed by the \, thus it gets confused on how to map the fields.Now let’s see if other rows contains the same problem:
grep '\\' provider.csv | wc -l
The command searches for the
\ character in the file and counts the lines which contain the pattern that we are looking for. The result is 70.There are many ways to fix this problem. Below, I replace the
\ with / and load it into a new file.tr "\\" "/" < provider.csv > provider_clean.csv
Now let’s try to reload the CSV file again. This time I am loading the file from Python client.
def create_provider_node(file, g):
query = '''
USING PERIODIC COMMIT 1000
LOAD CSV FROM {file} AS col
CREATE (pd:Provider {npi: col[0], entityType: col[1], address: col[20]+col[21], city: col[22], state: col[23], zip: col[24], country: col[25]})
FOREACH (row in CASE WHEN col[1]='1' THEN [1] else [] END | SET pd.firstName=col[6], pd.lastName = col[5], pd.credential= col[10], pd.gender = col[41])
FOREACH (row in CASE WHEN col[1]='2' THEN [1] else [] END | SET pd.orgName=col[4])
'''
index1 = '''
CREATE INDEX ON: Provider(npi)
'''
g.run(index1)
return g.run(query, file = file)
pw = os.environ.get('NEO4J_PASS')
g = Graph("https://localhost:7474/", password=pw)
tx = g.begin()
file = 'file:///provider_clean.csv'
create_provider_node(file, g)
By using periodic commit, you can set up a number of transactions to be committed. It helps to prevent from using large amount of memory when loading large CSV files.
4. Conclusion
Now, I have successfully loaded the healthcare provider data into Neo4j. The process of loading drug, drug manufacturer and prescription data are very similar. I also created the relationship
WRITES for the nodes :Provider and :Prescription based on the NPI information contained in both files. By now, all the data is stored in the graph database.Let’s take a look at the healthcare graph data model again:
I hope you find this blog post helpful. Sometimes cleaning large CSV files can be tricky, but using the command line to manipulate the files can make the work go faster. In the next blog post, I will show you how to link data when you have limited resources. Specifically, I will demonstrate how I created the relationship
(:Prescription)-[:PRESCRIBE]->(:Drug) and (:Drug Firm)-[BRANDS]->(:Drug). Stay tuned, and I’ll see you soon!Ready to dig in and get started with graph technology? Click below to download this free ebook, Learning Neo4j and catch up to speed with the world’s leading graph database.
Catch up with the rest of the Neo4j in healthcare series: