BioCypher: Unifying Framework for Biomedical Knowledge Graphs

Senior Developer Marketing Manager
3 min read
Sebastian Lobentanzer joined me to talk about how to make life easier for biotech experts and researchers by making the creation of knowledge graphs from various sources easier.
To watch the full episode, scroll to the end of this blog post.

The Creation of Knowledge Graphs
Recently, at Neo4j GraphSummit Munich I heard Katariina Kari talk about how much it matters to spend time upfront to get your semantic taxonomies in order before you set out to create a knowledge graph.
If this initial step is not built on stable grounds the rest of the process might fail and you will not achieve your goals.
Sebastian agrees with this concern and says that often teams don’t have the bandwidth to build their own processes and therefore rely on what is already there even though that might not be 100% aligned with their goals, but is deemed “good enough”.
This was the big motivation behind building BioCypher: making it easier to create a knowledge graph out of existing resources in combination with their own data and making that process more flexible at the same time.
BioCypher
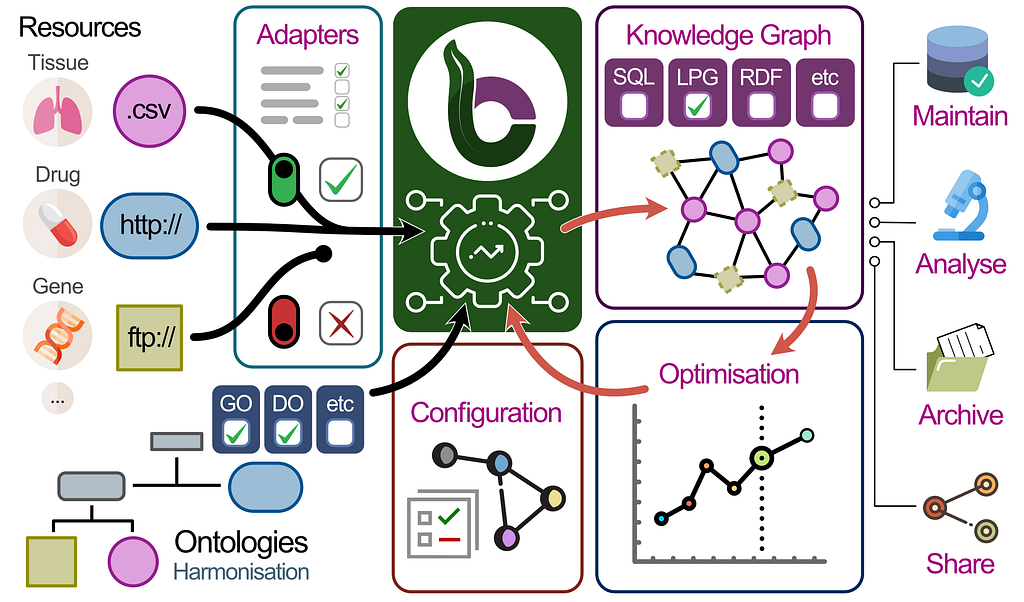
BioCypher tackles this by combining three main areas:
- Resources: Piping any kind of data into the framework via ETL-like processes. Through adapters, the data selected can be highly customized and helps the user define what information they need on the Node/Edge level (think Graph Modelling). So the user loads only the data they need and the pipeline already puts it in the right place. There are many adapters for all kinds of sources, e.g. Omnipath.
- Ontologies: Expertly curated information is mapped with the data. Additionally, the user can select which ontologies they want to use and link different ontologies together to fit their needs. In the future, it is planned to even allow merging ontologies.
- Output: The combined data is then exported into Property Graph, SQL or RDF for the users to then work with the data. The schema can also be shared so that other users can easily replicate it.
Using BioCypher
To conclude the episode, Sebastian presented a quick demo of BioCypher by loading a Metagraph into Neo4j.
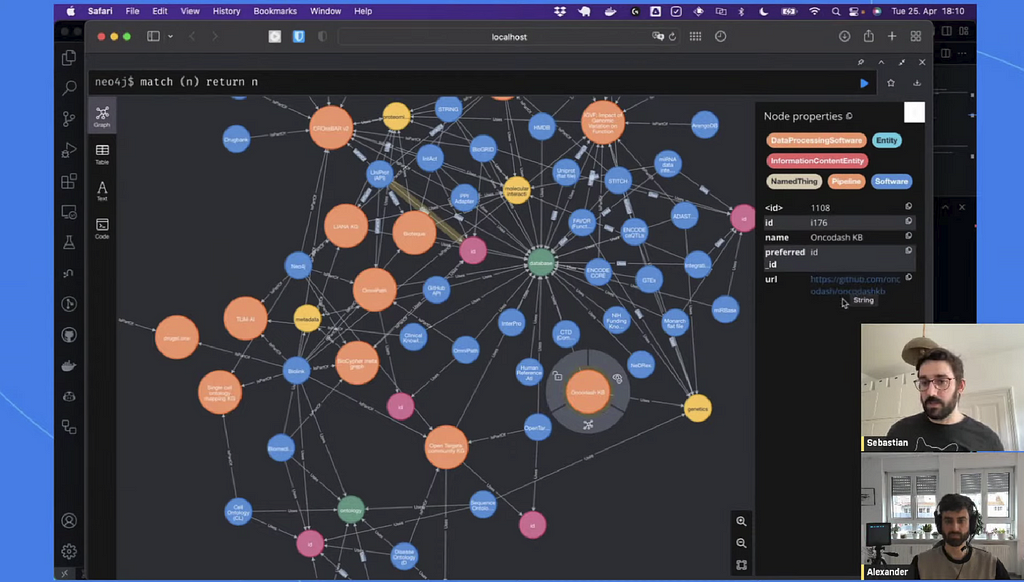
The team behind BioCypher is making this approach as robust as possible with unit testing and giving users verbose feedback on the structure of their knowledge graphs or highlighting duplicates.
For the future, they want to extend workflows per use case, ontology reasoning, improve performance and develop a more intuitive user interface (maybe even a GUI).
Help from interested members of the community is always welcome!
Watch the Full Episode
Interesting Links
BioCypher https://biocypher.org/
Github https://github.com/biocypher
Paper: https://arxiv.org/abs/2212.13543
Sebastian Lobentanzer https://twitter.com/slobentanzer
Graphs4Good: https://development.neo4j.dev/graphs4good/
HealthECCO: https://healthecco.org/
Omnipath: https://omnipathdb.org/
Going Meta: https://development.neo4j.dev/video/going-meta-a-series-on-graphs-semantics-and-knowledge/
BioCypher: Unifying Framework for Biomedical Knowledge Graphs was originally published in Neo4j Developer Blog on Medium, where people are continuing the conversation by highlighting and responding to this story.