BAML is a language created by Boundary for generating clean, structured output from unstructured data. Neo4j is a graph database for storing data as graphs — nodes and the relationships between them.
In this article, I’ll walk through how I extended one of BAML’s sample projects to convert web pages into graph representations to quickly populate a Neo4j instance.
Requirements
Workflow
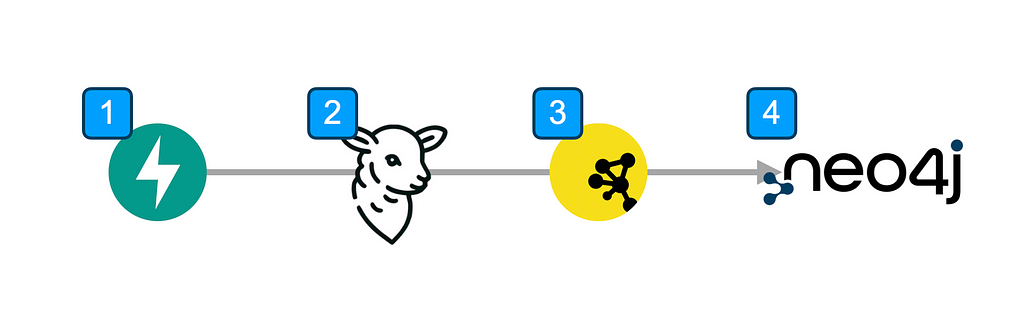
1. FastAPI
Boundary has sample code for running a simple FastAPI server using BAML to extract data from a hard-coded resume. FastAPI is a great framework that includes an interactive docs page that allows you to test endpoints right from a browser.
I originally forked this and added an experimental endpoint to extract entities and relationships from web-page content. This endpoint takes a list of URL strings as an argument.
The underlying function will then call BAML functions to extract data from the web pages as structured JSON objects uploaded to Neo4j.
@app.post("/url_to_graph")
async def extract_url_content(urls: list[str]):
"""General purpose conversion of contents from a list of urls to a graph"""
# 1. Prep html to text conversion
h = html2text.HTML2Text()
h.ignore_links = False
# 2. Extract text from each url
markdown_contents = []
for url in urls:
try:
response = requests.get(url)
response.raise_for_status()
html_content = response.text
markdown_content = h.handle(html_content)
markdown_contents.append(markdown_content)
except Exception as e:
markdown_contents.append(f"Error processing {url}: {str(e)}")
# 3. Composite the text
combined_markdown = "nn".join(markdown_contents)
# 4. Run BAML to get a Cytoscape graph JSON representation of text
json_output = b.GenerateCytoscapeGraph(combined_markdown)
json_str = str(json_output.model_dump_json())
json_dict = json.loads(json_str)
# 5. Upload the JSON data to Neo4j
finished = upload_cytoscape_to_neo4j(json_dict)
return {"finished": finished}2. BAML
Structurally, when using BAML, two subfolders are added to a root project folder: baml_src and baml_client.
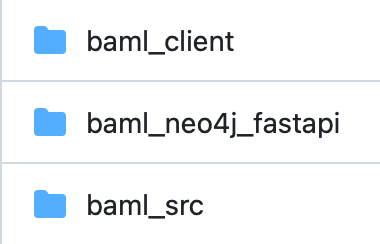
The contents of thebaml_client folder are auto-generated by either an active Visual Studio (VS) Code plugin or by manually running baml cli generate after updating any .baml files in the baml_src folder.
The baml_src folder requires main.baml and clients.baml files. The client.baml file includes specifications for foundation models to use, and main.baml specifies configuration options.
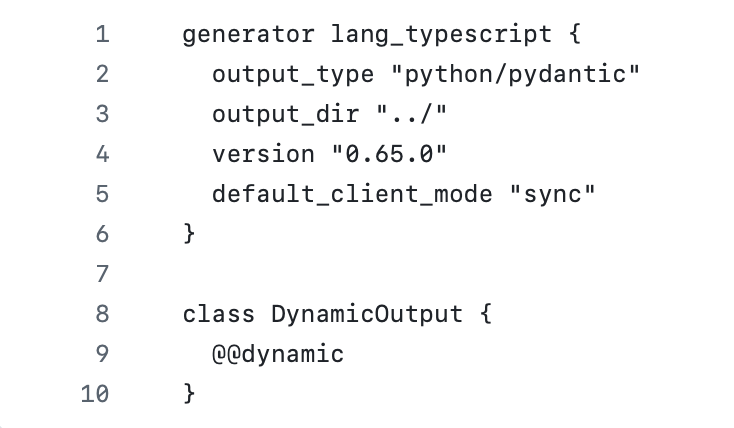
NOTE: The version number here should match the CLI version or the automatic baml-client content generation will stop working.
The .baml files you add will contain one or all three elements:
- Model specifications (required)
- Functions (required)
- Tests (optional)
BAML functions are where input-to-output definitions, client models to use, and LLM prompts can be specified. The following is the function the FastAPI endpoint calls to convert the HTML text into structured graph data:
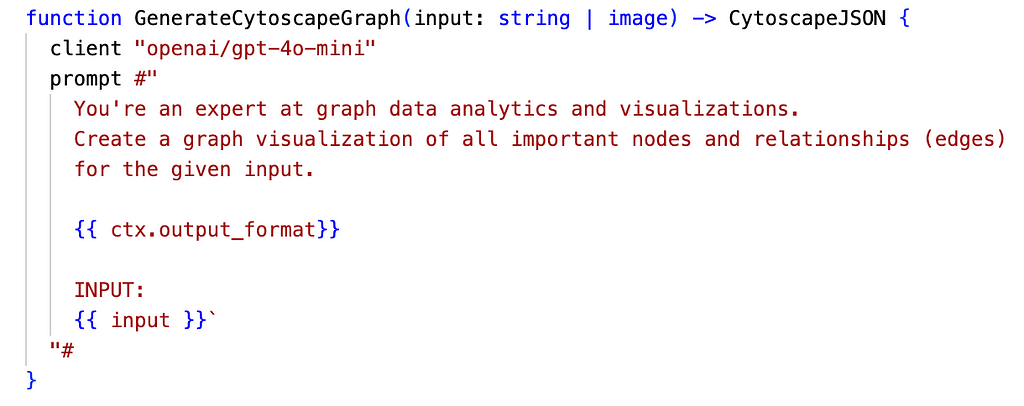
Model classes are defined in a similar but less verbose way to Pydantic, which is used under the hood. To get a structured JSON output like this:
{
"elements": {
"nodes": [
{
"data": {
"id": "neo4j_graph_database",
"name": "Neo4j Graph Database",
"label": "product",
"description": "Self or fully-managed, deploy anywhere"
}
},
{
"data": {
"id": "neo4j_auradb",
"name": "Neo4j AuraDB",
"label": "product",
"description": "Fully-managed graph database as a service"
}
},
],
"edges": [
{
"data": {
"id": "edge_1",
"source": "neo4j_graph_database",
"target": "neo4j_auradb",
"label": "RELATED_TO"
}
}
]
}
}The following model definition in BAML can be used:

NOTE: BAML class and function names are currently globally scoped, even when defined in different .baml files. Additionally, underscored prefixes in property names (example: _id) aren’t currently permitted.
3. Cytoscape
Cytoscape is a popular open source platform for visualizing graph or network data. The JSON data format it uses is simple yet flexible enough to support additional data besides the required id and label properties for nodes.
An example of this data was presented earlier with the BAML model description.
I used Cytoscape.js as the intermediary graph data format because Neo4j doesn’t have a preferred JSON format and because I’ll use Cytoscape to visual graph data in future demos.
4. Neo4j
Neo4j is one of the most popular graph databases available. Natively, it uses the Cypher query language to input, manipulate, and output graph data. As such, for this demo I used the officially supported Neo4j Bolt Python package to upload the Cytoscape-formatted JSON data in the cytoscape2neo4j.py file.
Walking through the key upload_cytoscape_to_neo4j function would warrant its own article. But in short, it loops through each node and relationship defined in the source JSON data and creates a Cypher query to create a node for each one specified and a relationship for each edge specified.
Using an Object Graph Mapper (OGM) package, such as Neomodel, or setting up a Apollo GraphQL server are alternate ways for creating this input bridge between JSON and Neo4j.
Running
All the code presented can be found at this public Github repo. To run it with Poetry, run the following commands from a command line:
poetry install
poetry run uvicorn baml_neo4j_fastapi.app:app - reload
Once running, the following interactive docs will be accessible from your browser at localhost:8000/docs:
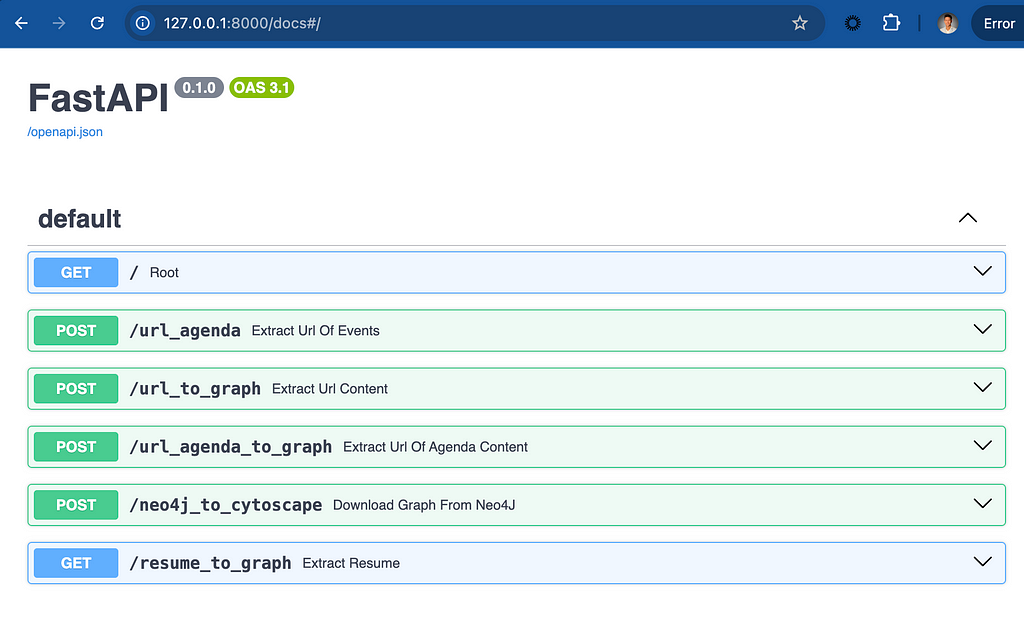
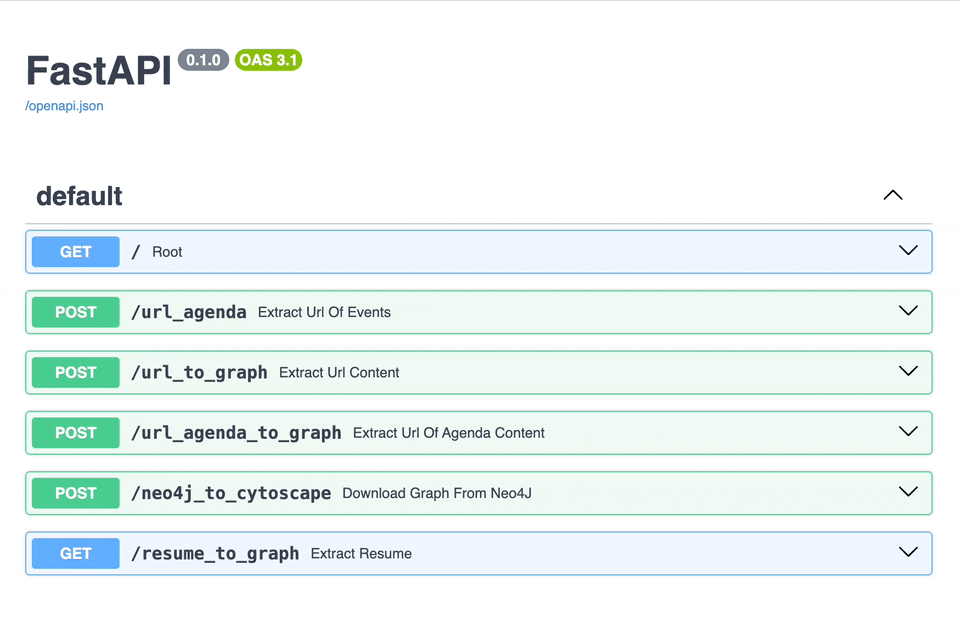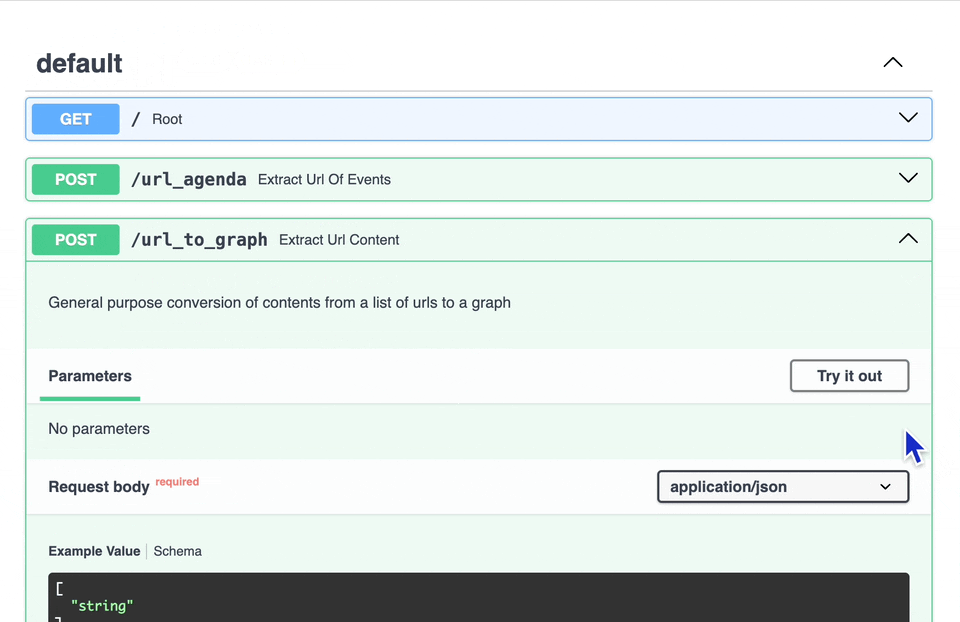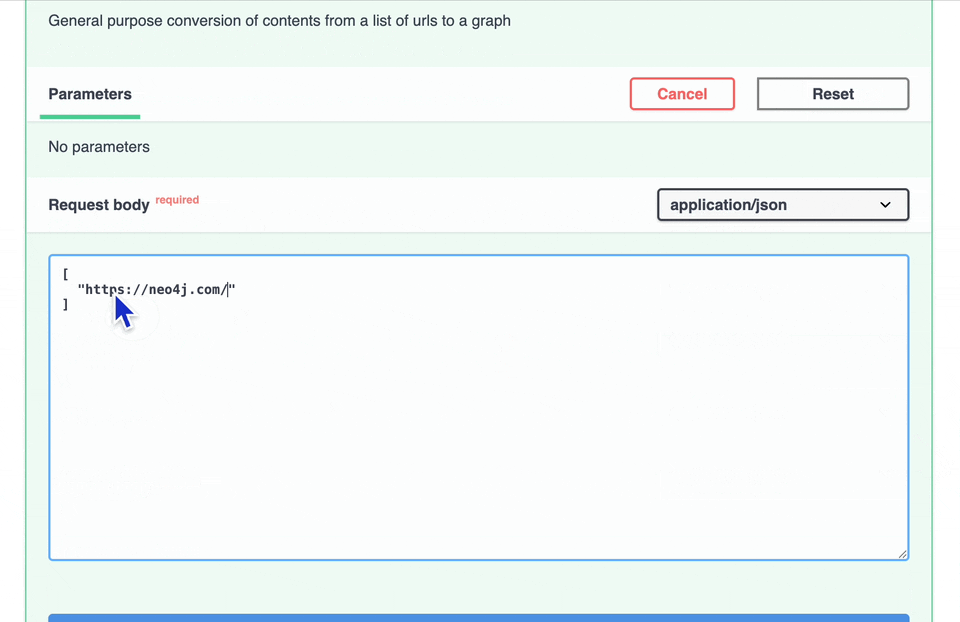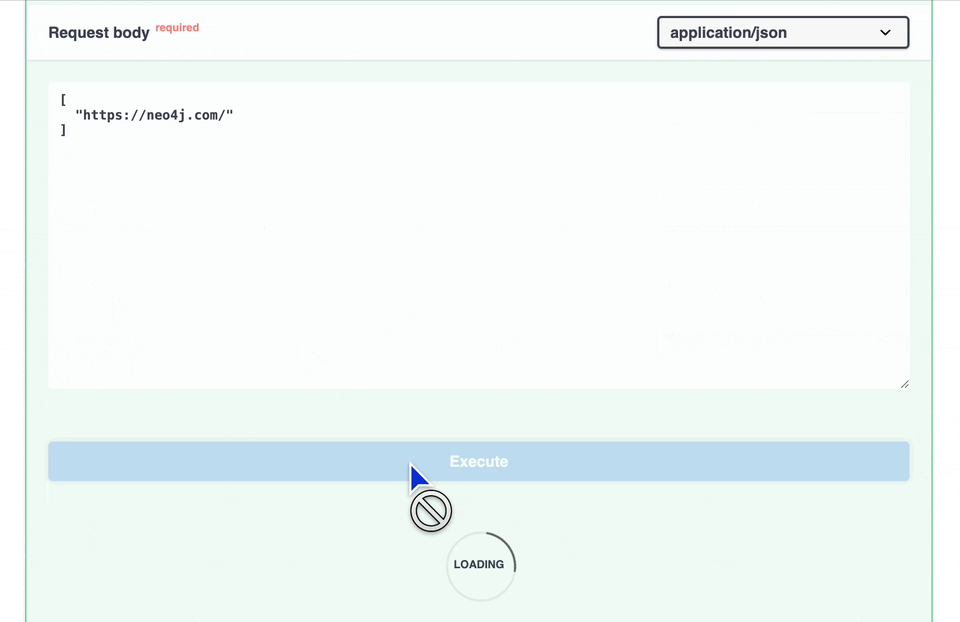
There are some other experimental endpoints, but the one discussed here is for the /url_to_graph option.
To test, open the endpoint details, click the Try It Out button, add one or more valid URLs, then click Execute.
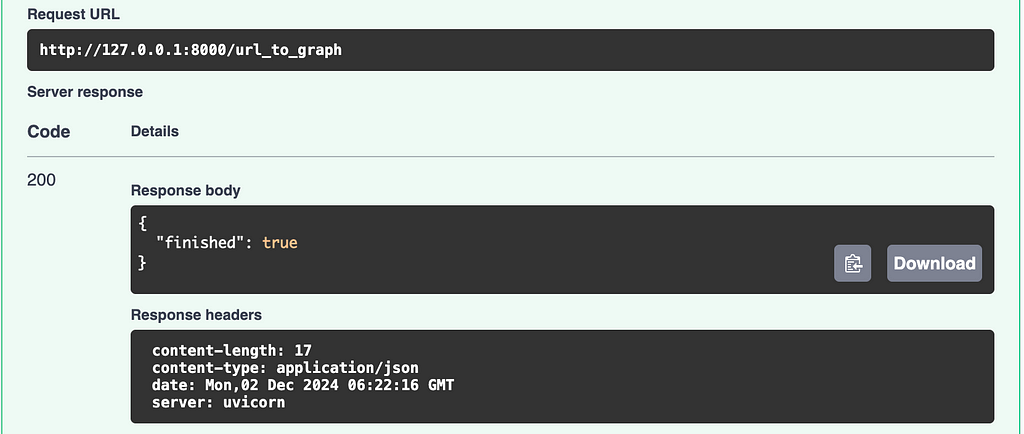
After a few moments, depending on how many and how large the source web pages are, you’ll hopefully get a finished:true response.
And in the terminal/console, you should see something like:
...
---Parsed Response (class CytoscapeJSON)---
{
"elements": {
"nodes": [
{
"data": {
"id": "neo4j_graph_database",
"name": "Neo4j Graph Database",
"label": "product",
"description": "Self or fully-managed, deploy anywhere"
}
},
{
"data": {
"id": "neo4j_auradb",
"name": "Neo4j AuraDB",
"label": "product",
"description": "Fully-managed graph database as a service"
}
},
{
"data": {
"id": "generative_ai",
"name": "Generative AI",
"label": "use_case",
"description": "Back your LLMs with a knowledge graph for better business AI"
}
}
],
"edges": [
{
"data": {
"id": "prod_use_case_1",
"source": "neo4j_graph_database",
"target": "generative_ai",
"label": "ENABLED_BY"
}
},
{
"data": {
"id": "prod_use_case_2",
"source": "neo4j_auradb",
"target": "generative_ai",
"label": "ENABLED_BY"
}
}
]
}
}
INFO: 127.0.0.1:63190 - "POST /url_to_graph HTTP/1.1" 200 OK
And in your Neo4j console, something like:
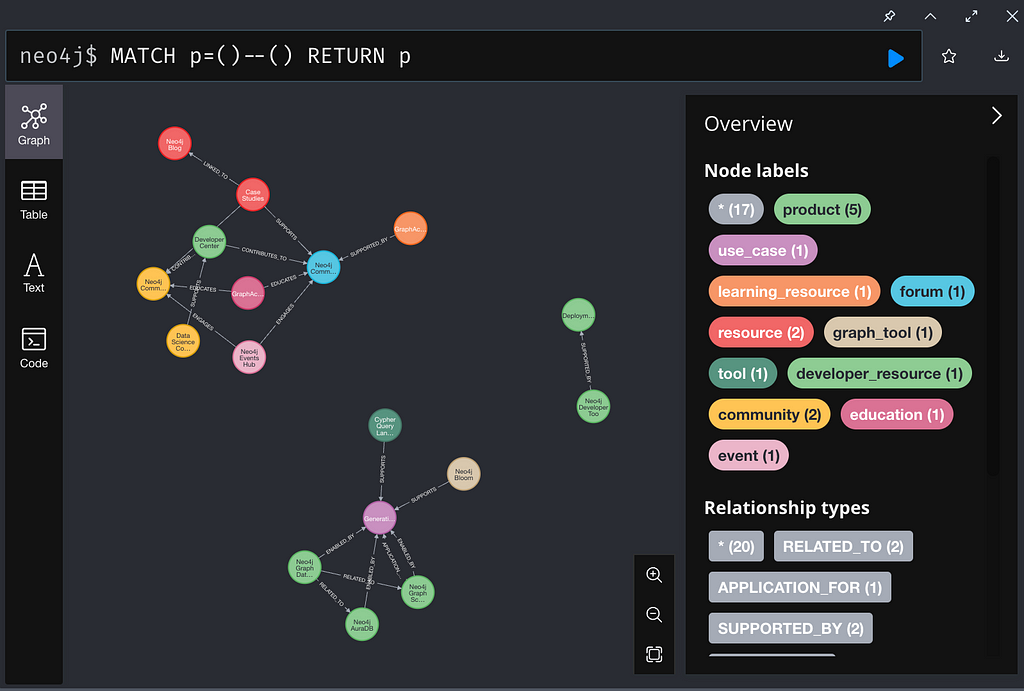
Next Steps
- Now that you have some graph data, you can start exploring various GraphRAG techniques to power GenAI applications.
- Learn more about BAML through their official docs.
- Learn more about using Cypher and Neo4j through GraphAcademy, a free online courses platform.
Unstructured-to-Structured Graph Data Using BAML to Neo4j was originally published in Neo4j Developer Blog on Medium, where people are continuing the conversation by highlighting and responding to this story.