GraphRAG Python Package: Accelerating GenAI With Knowledge Graphs

Data Science Product Specialist, Neo4j
11 min read
The GraphRAG Python package from Neo4j provides end-to-end workflows that take you from unstructured data to knowledge graph creation, knowledge graph retrieval, and full GraphRAG pipelines in one place. Whether you’re using Python to build knowledge assistants, search APIs, chatbots, or report generators, this package makes it easy to incorporate knowledge graphs to improve your retrieval-augmented generation (RAG) relevance, accuracy, and explainability.
In this post, we’ll show you how to go from zero to GraphRAG using the GraphRAG Python package and how you can use different knowledge graph retrieval patterns to get different behavior from your GenAI app.
GraphRAG: Adding Knowledge to GenAI
Combining knowledge graphs and RAG, GraphRAG helps solve common issues with large language models (LLMs), like hallucinations, while adding domain-specific context for improved quality and effectiveness over traditional RAG approaches. Knowledge graphs provide the contextual data LLMs need to reliably answer questions and serve as trusted agents in complex workflows. Unlike most RAG solutions, which only offer access to fragments of textual data, GraphRAG can integrate both structured and semi-structured information into the retrieval process.
The GraphRAG Python package will help you create knowledge graphs and easily implement knowledge graph data retrieval patterns that use combinations of graph traversals, query generation with text2Cypher, vector, and full-text search. Combined with additional tooling to support full RAG pipelines, this package allows you to seamlessly implement GraphRAG in your GenAI applications and workflows.
A Quick Overview: GraphRAG in a Few Lines of Code
Below are a few lines of code for a full end-to-end workflow. In the following sections, we’ll explore a more detailed version of this example and provide additional resources to kickstart your next GenAI breakthrough. You can find all the code to replicate in this GitHub repository.
import neo4j
from neo4j_graphrag.llm import OpenAILLM as LLM
from neo4j_graphrag.embeddings.openai import OpenAIEmbeddings as Embeddings
from neo4j_graphrag.experimental.pipeline.kg_builder import SimpleKGPipeline
from neo4j_graphrag.retrievers import VectorRetriever
from neo4j_graphrag.generation.graphrag import GraphRAG
neo4j_driver = neo4j.GraphDatabase.driver(NEO4J_URI,
auth=(NEO4J_USERNAME, NEO4J_PASSWORD))
ex_llm=LLM(
model_name="gpt-4o-mini",
model_params={
"response_format": {"type": "json_object"},
"temperature": 0
})
embedder = Embeddings()
# 1. Build KG and Store in Neo4j Database
kg_builder_pdf = SimpleKGPipeline(
llm=ex_llm,
driver=neo4j_driver,
embedder=embedder,
from_pdf=True
)
await kg_builder_pdf.run_async(file_path='precision-med-for-lupus.pdf')
# 2. KG Retriever
vector_retriever = VectorRetriever(
neo4j_driver,
index_name="text_embeddings",
embedder=embedder
)
# 3. GraphRAG Class
llm = LLM(model_name="gpt-4o")
rag = GraphRAG(llm=llm, retriever=vector_retriever)
# 4. Run
response = rag.search( "How is precision medicine applied to Lupus?")
print(response.answer)
#output
Precision medicine in systemic lupus erythematosus (SLE) is an evolving approach that aims to tailor treatment based on individual genetic, epigenetic, and pathophysiological characteristics…
0. Create a Neo4j Database
To begin our RAG example, we need to create a database to use for retrieval. You can quickly start a free Neo4j Graph Database using Neo4j AuraDB. You can use AuraDB Free or start an AuraDB Professional (Pro) free trial for higher ingestion and retrieval performance. The Pro instances have a bit more RAM; we recommend them for the best user experience.
Once you create an instance, you can download and save the credentials to use in the following code.
1. Build a Knowledge Graph
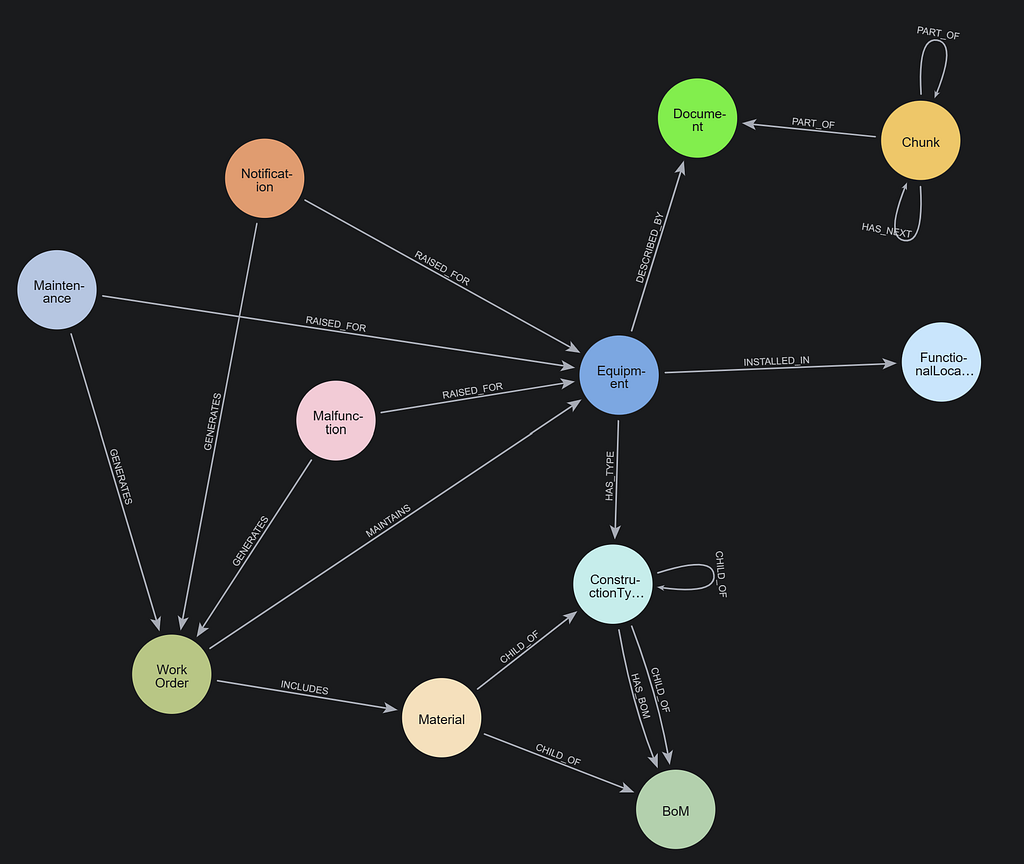
We’ll transform documents into a knowledge graph and store it in our Neo4j database. We will demonstrate with a few PDF documents containing medical research papers on Lupus. These documents contain specialized, domain-specific information, and a knowledge graph can enhance AI use cases by organizing this complex clinical data more effectively. Below is a sample of the graph that we will create. It contains a few key node types (a.k.a. node labels):
Document: metadata for document sourcesChunk: text chunks from the documents with embeddings to power vector retrieval__Entity__: Entities extracted from the text chunks
Notice the relationships between entities and how they create connect paths between chunks and documents. This is an advantage of GraphRAG, and we will see how it becomes important later.
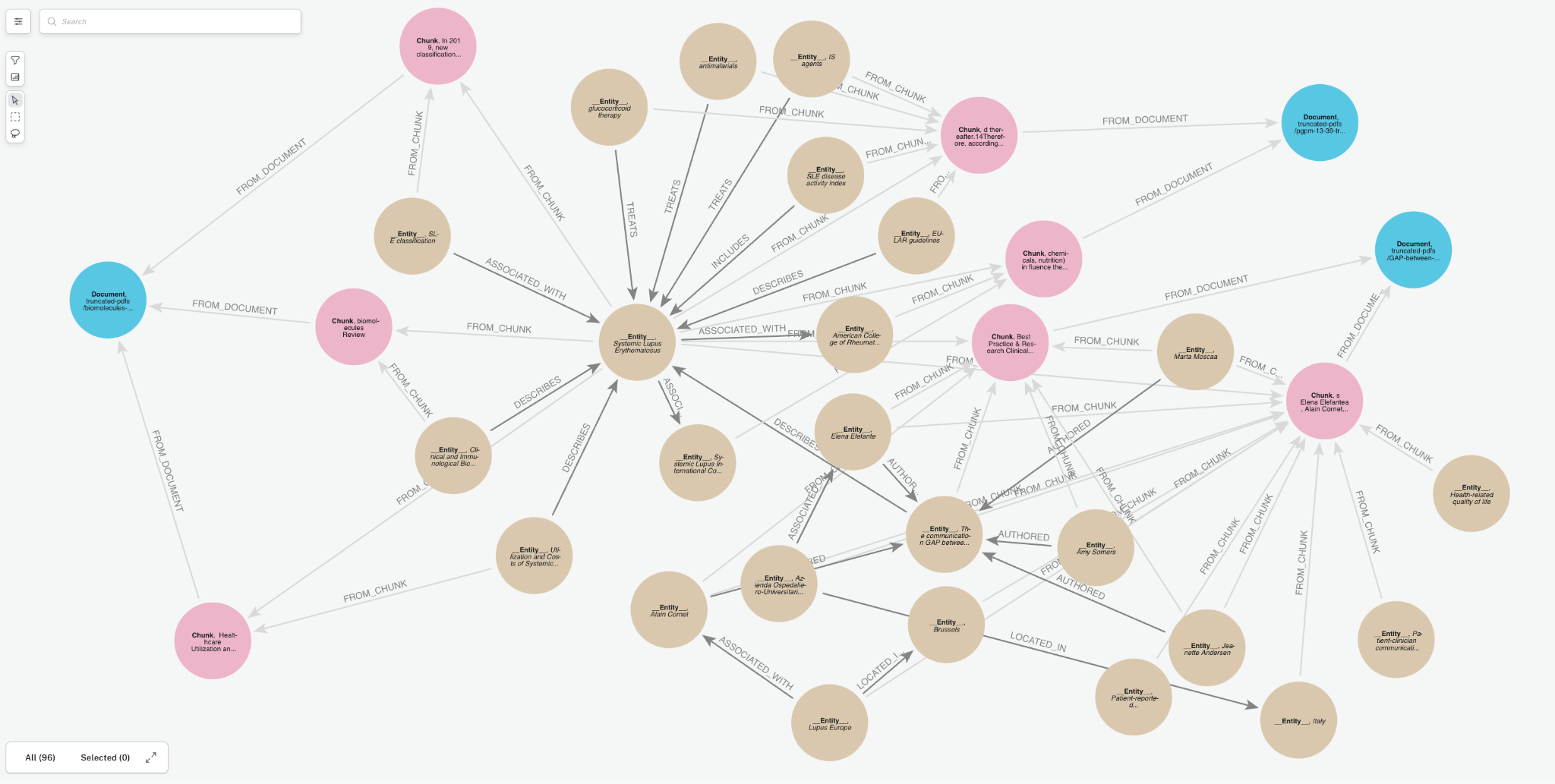
Creating a knowledge graph with the GraphRAG Python package is pretty simple—even if you’re not a Neo4j expert.
The SimpleKGPipeline class allows you to automatically build a knowledge graph with a few key inputs, including
- a driver to connect to Neo4j,
- an LLM for entity extraction, and
- an embedding model to create vectors on text chunks for similarity search.
Neo4j Driver
The Neo4j driver allows you to connect and perform read and write transactions with the database. You can obtain the URI, username, and password variables from when you created the database. If you created your database on AuraDB, they are in the file you downloaded.
import neo4j
neo4j_driver = neo4j.GraphDatabase.driver(NEO4J_URI,
auth=(NEO4J_USERNAME, NEO4J_PASSWORD))
LLM & Embedding Model
In this case, we will use OpenAI GPT-4o-mini for convenience. It is a fast and low-cost model. The GraphRAG Python package supports any LLM model, including models from OpenAI, Google VertexAI, Anthropic, Cohere, Azure OpenAI, local Ollama models, and any chat model that works with LangChain. You can also implement a custom interface for any other LLM.
Likewise, we will use OpenAI’s default text-embedding-ada-002 for the embedding model, but you can use other embedders from different providers.
import neo4j
from neo4j_graphrag.llm import OpenAILLM
from neo4j_graphrag.embeddings.openai import OpenAIEmbeddings
llm=OpenAILLM(
model_name="gpt-4o-mini",
model_params={
"response_format": {"type": "json_object"}, # use json_object formatting for best results
"temperature": 0 # turning temperature down for more deterministic results
}
)
#create text embedder
embedder = OpenAIEmbeddings()
Optional Inputs: Schema & Prompt Template
While not required, adding a graph schema is highly recommended for improving knowledge graph quality. It provides guidance for the node and relationship types to create during entity extraction.
Pro-tip: If you are still deciding what schema to use, try building a graph without a schema first and examine the most common node and relationship types created as a starting point.
For our graph schema, we will define entities (a.k.a. node labels) and relations that we want to extract. While we won’t use it in this simple example, there is also an optional potential_schema argument, which can guide which relationships should connect to which nodes.
basic_node_labels = ["Object", "Entity", "Group", "Person", "Organization", "Place"]
academic_node_labels = ["ArticleOrPaper", "PublicationOrJournal"]
medical_node_labels = ["Anatomy", "BiologicalProcess", "Cell", "CellularComponent",
"CellType", "Condition", "Disease", "Drug",
"EffectOrPhenotype", "Exposure", "GeneOrProtein", "Molecule",
"MolecularFunction", "Pathway"]
node_labels = basic_node_labels + academic_node_labels + medical_node_labels
# define relationship types
rel_types = ["ACTIVATES", "AFFECTS", "ASSESSES", "ASSOCIATED_WITH", "AUTHORED",
"BIOMARKER_FOR", …]
We will also be adding a custom prompt for entity extraction. While the GraphRAG Python package has an internal default prompt, engineering a prompt closer to your use case often helps create a more applicable knowledge graph. The prompt below was created with a bit of experimentation. Be sure to follow the same general format as the default prompt.
prompt_template = '''
You are a medical researcher tasks with extracting information from papers
and structuring it in a property graph to inform further medical and research Q&A.
Extract the entities (nodes) and specify their type from the following Input text.
Also extract the relationships between these nodes. the relationship direction goes from the start node to the end node.
Return result as JSON using the following format:
{{"nodes": [ {{"id": "0", "label": "the type of entity", "properties": {{"name": "name of entity" }} }}],
"relationships": [{{"type": "TYPE_OF_RELATIONSHIP", "start_node_id": "0", "end_node_id": "1", "properties": {{"details": "Description of the relationship"}} }}] }}
- Use only the information from the Input text. Do not add any additional information.
- If the input text is empty, return empty Json.
- Make sure to create as many nodes and relationships as needed to offer rich medical context for further research.
- An AI knowledge assistant must be able to read this graph and immediately understand the context to inform detailed research questions.
- Multiple documents will be ingested from different sources and we are using this property graph to connect information, so make sure entity types are fairly general.
Use only fhe following nodes and relationships (if provided):
{schema}
Assign a unique ID (string) to each node, and reuse it to define relationships.
Do respect the source and target node types for relationship and
the relationship direction.
Do not return any additional information other than the JSON in it.
Examples:
{examples}
Input text:
{text}
'''
Creating the SimpleKGPipeline
Create the SimpleKGPipeline using the constructor below:
from neo4j_graphrag.experimental.components.text_splitters.fixed_size_splitter import FixedSizeSplitter from neo4j_graphrag.experimental.pipeline.kg_builder import SimpleKGPipeline kg_builder_pdf = SimpleKGPipeline( llm=ex_llm, driver=driver, text_splitter=FixedSizeSplitter(chunk_size=500, chunk_overlap=100), embedder=embedder, entities=node_labels, relations=rel_types, prompt_template=prompt_template, from_pdf=True )
Other optional inputs for the SimpleKGPipeline include document loader and knowledge graph writer components. The full documentation is here.
Running the Knowledge Graph Builder
You can run the knowledge graph builder with the run_async method. We are going to iterate through 3 PDFs below.
pdf_file_paths = ['truncated-pdfs/biomolecules-11-00928-v2-trunc.pdf',
'truncated-pdfs/GAP-between-patients-and-clinicians_2023_Best-Practice-trunc.pdf',
'truncated-pdfs/pgpm-13-39-trunc.pdf']
for path in pdf_file_paths:
print(f"Processing : {path}")
pdf_result = await kg_builder_pdf.run_async(file_path=path)
print(f"Result: {pdf_result}")
Once complete, you can explore the resulting knowledge graph. The Unified Console provides a great interface for this.
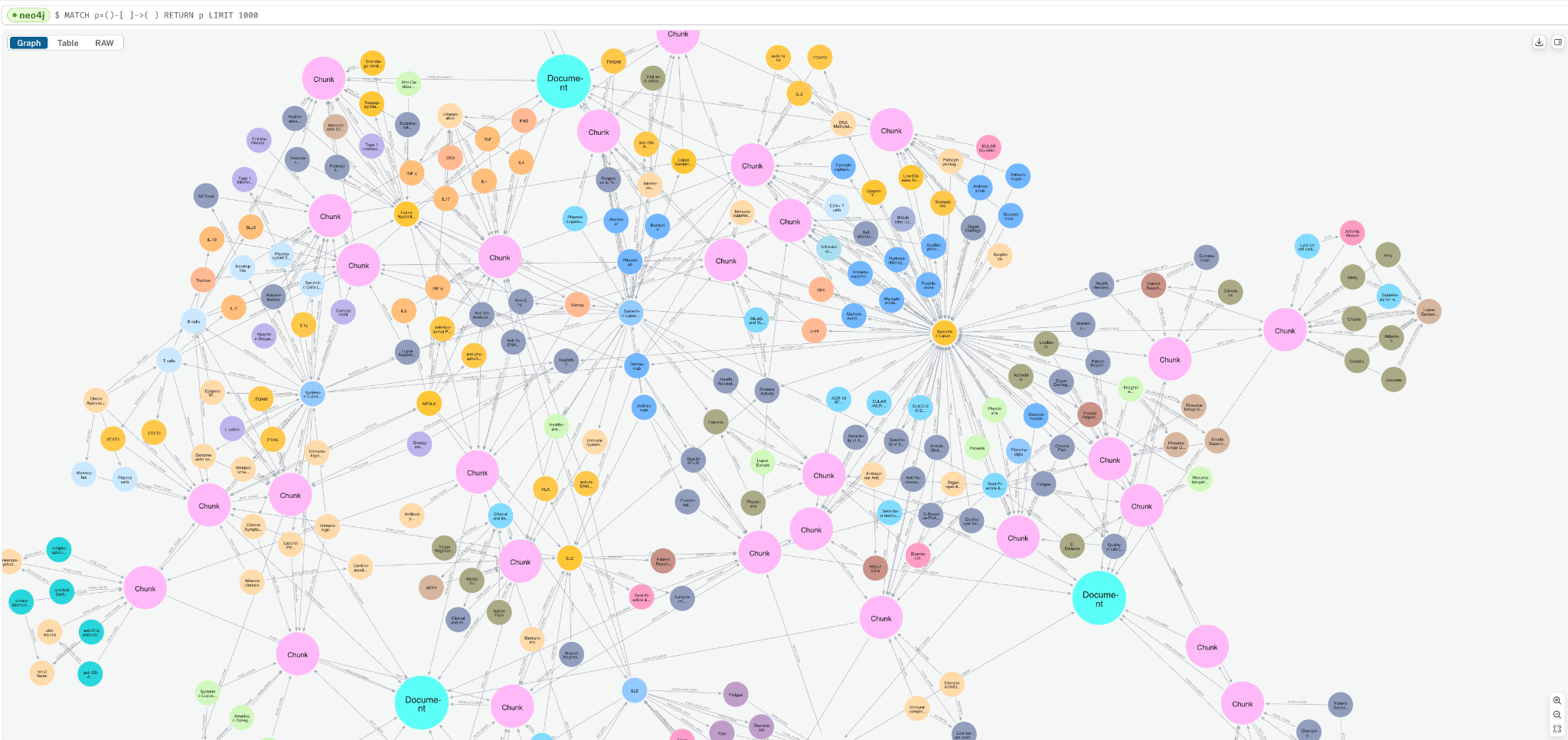
Go to the Query tab and enter the below query to see a sample of the graph.
MATCH p=()-->() RETURN p LIMIT 1000;
You can see how the Document, Chunk, and __Entity__ nodes are all connected together.
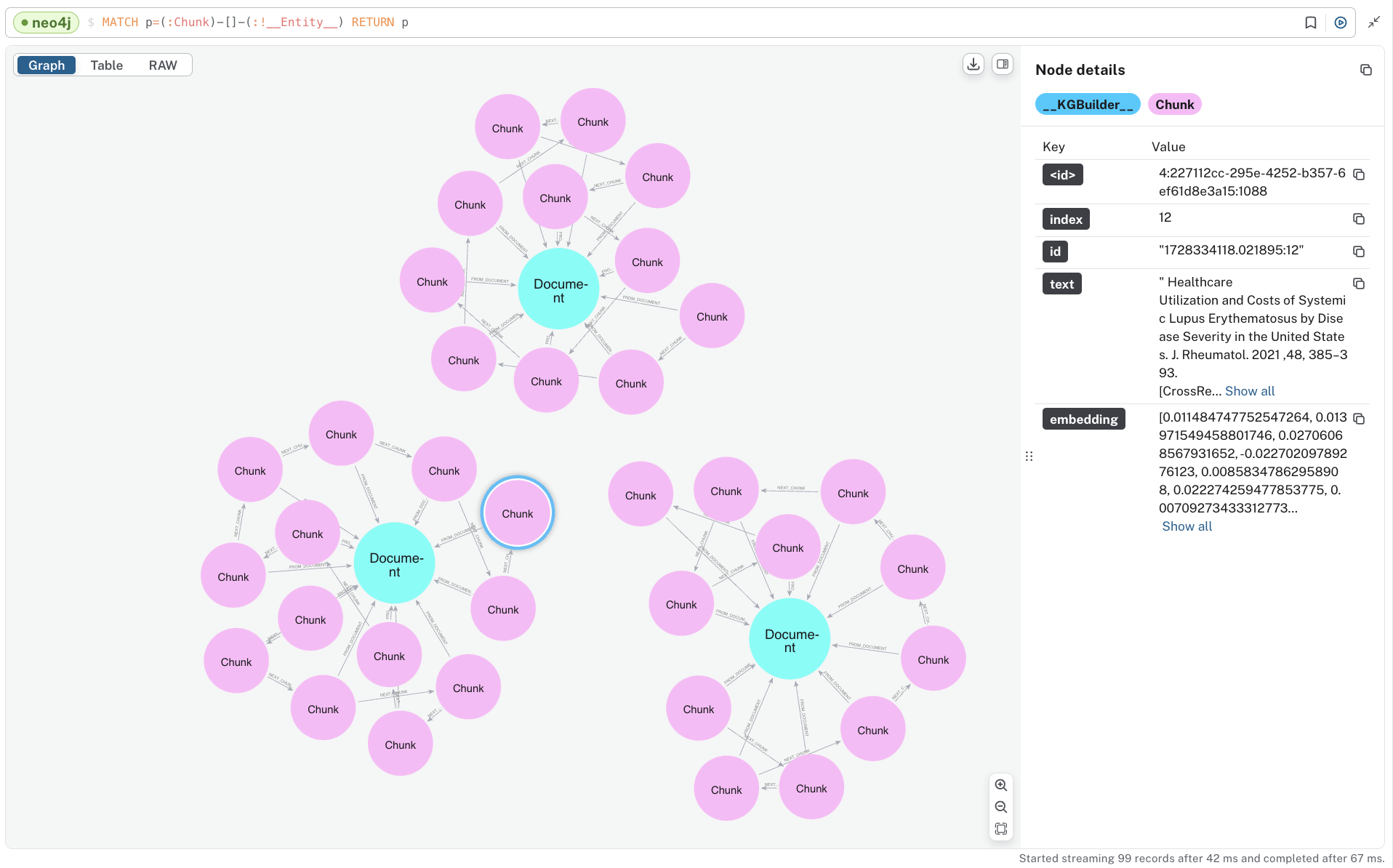
To see the “lexical” portion of the graph containing Document and Chunk nodes, run the following.
MATCH p=(:Chunk)--(:!__Entity__) RETURN p;
Note that these are disconnected components, one for each document we ingested. You can also see the embeddings that have been added to all chunks.
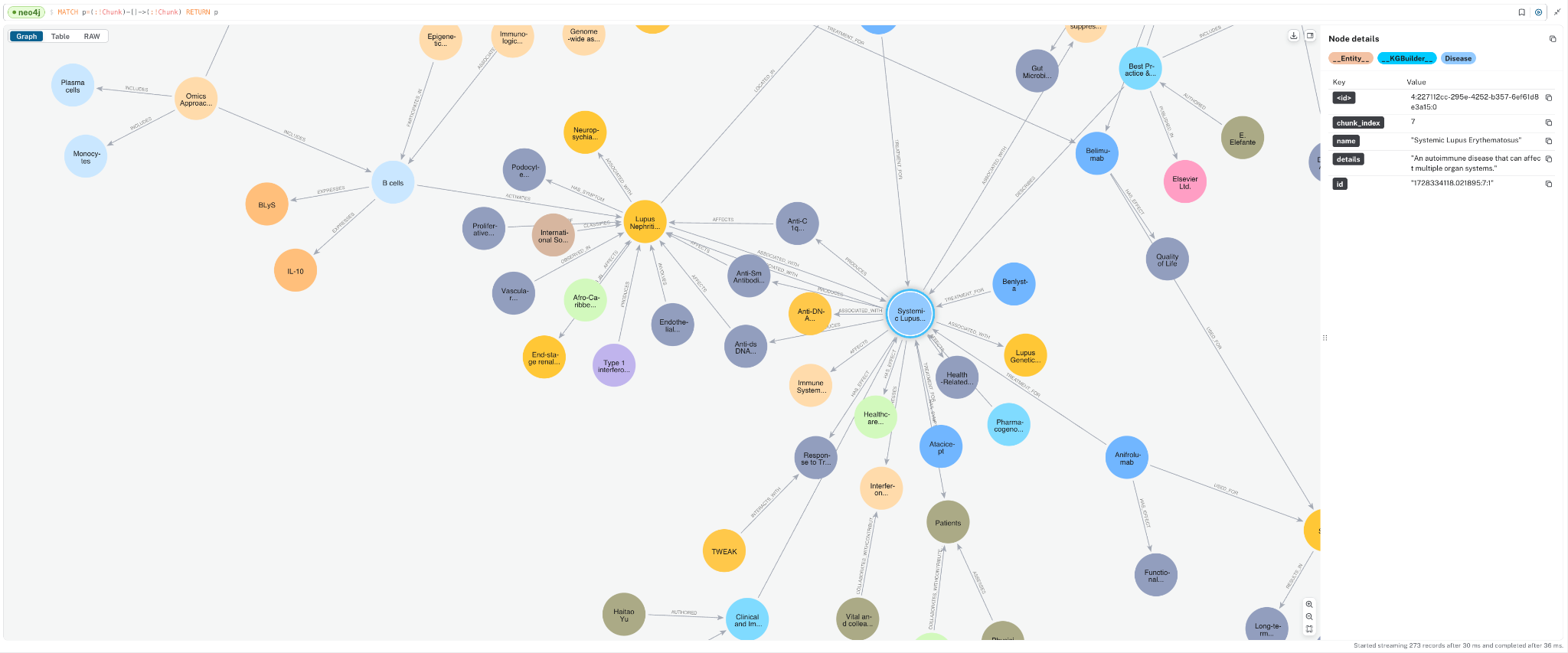
To look at just the domain graph of __Entity__ nodes, you can run the following:
MATCH p=(:!Chunk)-->(:!Chunk) RETURN p;
You will see how different concepts have been extracted and how they connect to one another. This domain graph connects information between the documents.
The Developer’s Guide to GraphRAG
Combine a knowledge graph with RAG to build a contextual, explainable GenAI app. Get started by learning the three main patterns.
A Note on Custom & Detailed Knowledge Graph Building
Under the Hood, the SimpleKGPipeline runs the components listed below. The GraphRAG package provides a lower-level pipeline API, allowing you to customize the knowledge graph-building process to a great degree. For further details, see this documentation.
- Document Parser: extract text from documents, such as PDFs.
- Text Splitter: split text into smaller pieces manageable by the LLM context window (token limit).
- Chunk Embedder: compute the text embeddings for each chunk
- Schema Builder: provide a schema to ground the LLM entity extraction for an accurate and easily navigable knowledge graph.
- Entity & Relation Extractor: extract relevant entities and relations from the text
- Knowledge Graph Writer: save the identified entities and relations to the KG
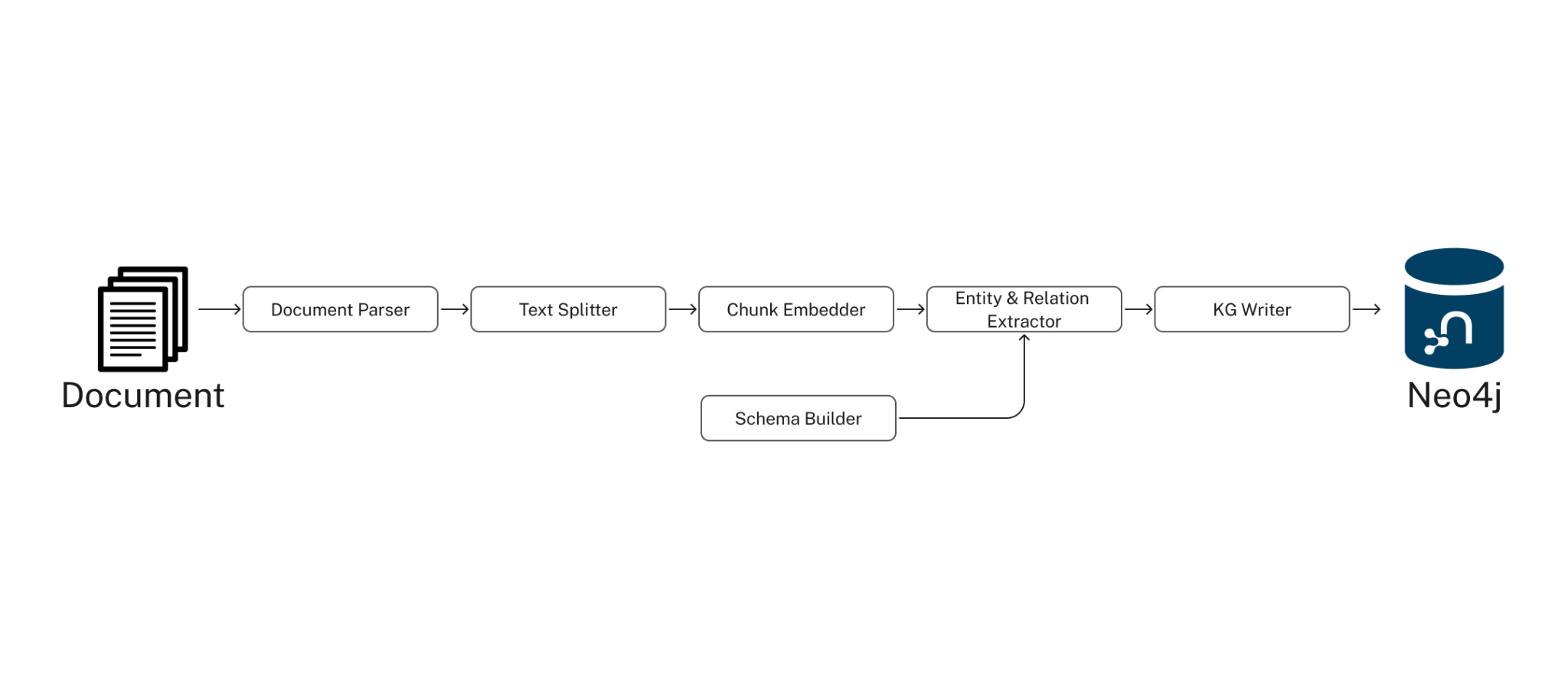
2. Retrieve Data From Your Knowledge Graph
The GraphRAG Python package provides multiple classes for retrieving data from your knowledge graph, including:
- Vector Retriever: performs similarity searches using vector embeddings
- Vector Cypher Retriever: combines vector search with retrieval queries in Cypher, Neo4j’s Graph Query language, to traverse the graph and incorporate additional nodes and relationships.
- Hybrid Retriever: Combines vector and full-text search.
- Hybrid Cypher Retriever: Combines vector and full-text search with Cypher retrieval queries for additional graph traversal.
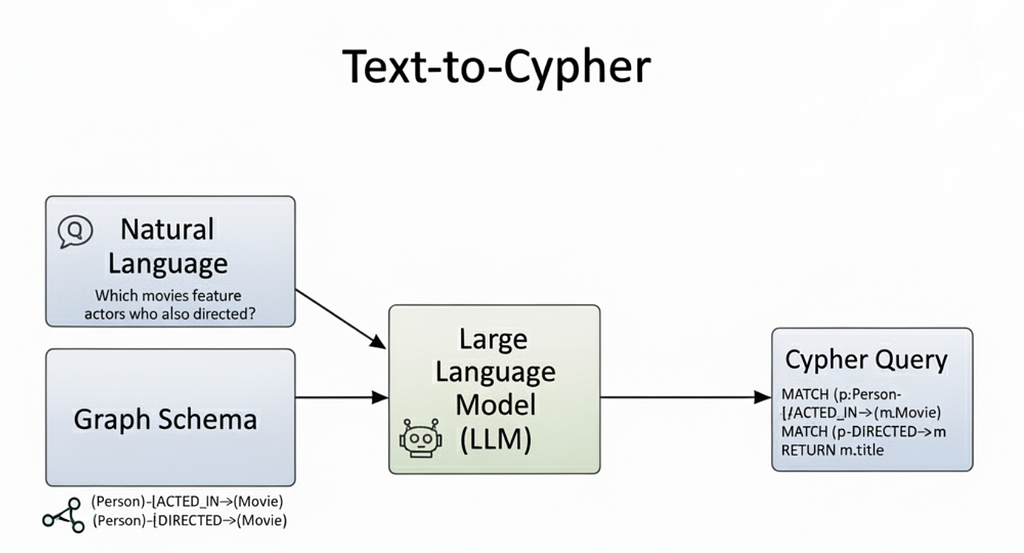
- Text2Cypher: converts natural language queries into Cypher queries to run against Neo4j.
- Weaviate & Pinecone Neo4j Retriever: Allows you to search vectors stored in Weaviate or Pinecone and connect them to nodes in Neo4j using external id properties.
- Custom Retriever: allows for tailored retrieval methods based on specific needs.
These retrievers enable you to implement diverse data retrieval patterns, boosting the relevance and accuracy of your RAG pipelines.
Let’s explore a couple of them on the graph we just created.
Vector Retriever
The Vector Retriever uses Approximate Nearest Neighbor (ANN) vector search to retrieve data from your knowledge graph.
We can create a vector index in Neo4j to allow this retriever to pull back information from Chunk nodes.
from neo4j_graphrag.indexes import create_vector_index
create_vector_index(driver, name="text_embeddings", label="Chunk",
embedding_property="embedding", dimensions=1536, similarity_fn="cosine")
You can then instantiate the vector retriever using the code below:
from neo4j_graphrag.retrievers import VectorRetriever vector_retriever = VectorRetriever( driver, index_name="text_embeddings", embedder=embedder, return_properties=["text"], )
Now, let’s run this retriever with a simple prompt. The retriever can pull back text chunks and vector similarity scores, providing some useful context for answering questions in RAG.
import json
vector_res = vector_retriever.get_search_results(query_text = "How is precision medicine applied to Lupus?",
top_k=3)
for i in vector_res.records: print("====n" + json.dumps(i.data(), indent=4))
# output text chunks
====
{
"node": {
"text": "precise and systematic fashion as suggested here.nFuture care will involve molecular diagnostics throughoutnthe patient timecourse to drive the least toxic combinationnof therapies. Recent evidence suggests a paradigm shift isnon the way but it is hard to predict how fast it will come.nDisclosurenThe authors report no con ufb02icts of interest in this work.nReferencesn1. Lisnevskaia L, Murphy G, Isenberg DA. Systemic lupusnerythematosus. Lancet .2014 ;384:1878 u20131888. doi:10.1016/S0140-n6736(14)60128"
},
"score": 0.9368438720703125
}
====
{
"node": {
"text": "d IS agents.nPrecision medicine consists of a tailored approach toneach patient, based on genetic and epigenetic singularities,nwhich in ufb02uence disease pathophysiology and drugnresponse. Precision medicine in SLE is trying to addressnthe need to assess SLE patients optimally, predict diseasencourse and treatment response at diagnosis. Ideally everynpatient would undergo an initial evaluation that wouldnproufb01le his/her disease, assessing the main pathophysiolo-ngic pathway through biomarkers, ther"
},
"score": 0.935699462890625
}
====
{
"node": {
"text": "REVIEWnT owards Precision Medicine in Systemic LupusnErythematosusnThis article was published in the following Dove Press journal:nPharmacogenomics and Personalized MedicinenElliott Lever1nMarta R Alves2nDavid A Isenberg1n1Centre for Rheumatology, Division ofnMedicine, University College HospitalnLondon, London, UK;2Internal Medicine,nDepartment of Medicine, CentronHospitalar do Porto, Porto, PortugalAbstract: Systemic lupus erythematosus (SLE) is a remarkable condition characterised byndiversit"
},
"score": 0.9312744140625
}
Vector Cypher Retriever
Another useful retriever to explore is the Vector Cypher Retriever, which lets you use Cypher, Neo4j’s graph query language, to incorporate graph traversal logic after retrieving an initial set of nodes with vector search. Below we create a retriever to obtain Chunk nodes via vector search, then traversing out on entities up to 3 hops out.
from neo4j_graphrag.retrievers import VectorCypherRetriever
vc_retriever = VectorCypherRetriever(
driver,
index_name="text_embeddings",
embedder=embedder,
retrieval_query="""
//1) Go out 2-3 hops in the entity graph and get relationships
WITH node AS chunk
MATCH (chunk)<-[:FROM_CHUNK]-()-[relList:!FROM_CHUNK]-{1,2}()
UNWIND relList AS rel
//2) collect relationships and text chunks
WITH collect(DISTINCT chunk) AS chunks,
collect(DISTINCT rel) AS rels
//3) format and return context
RETURN '=== text ===n' + apoc.text.join([c in chunks | c.text], 'n---n') + 'nn=== kg_rels ===n' +
apoc.text.join([r in rels | startNode(r).name + ' - ' + type(r) + '(' + coalesce(r.details, '') + ')' + ' -> ' + endNode(r).name ], 'n---n') AS info
"""
)
Submitting the same prompt, here’s the context we get back:
vc_res = vc_retriever.get_search_results(query_text = "How is precision medicine applied to Lupus?", top_k=3)
# print output
kg_rel_pos = vc_res.records[0]['info'].find('nn=== kg_rels ===n')
print("# Text Chunk Context:")
print(vc_res.records[0]['info'][:kg_rel_pos])
print("# KG Context From Relationships:")
print(vc_res.records[0]['info'][kg_rel_pos:])
# output
# Text Chunk Context:
=== text ===
precise and systematic fashion as suggested here.
Future care will involve molecular diagnostics throughout
the patient timecourse to drive the least toxic combination
of therapies. Recent evidence suggests a paradigm shift is
on the way but it is hard to predict how fast it will come.
Disclosure
The authors report no con flicts of interest in this work.
References
1. Lisnevskaia L, Murphy G, Isenberg DA. Systemic lupus
erythematosus. Lancet .2014 ;384:1878 –1888. doi:10.1016/S0140-
6736(14)60128
---
d IS agents.
Precision medicine consists of a tailored approach to
each patient, based on genetic and epigenetic singularities,
which in fluence disease pathophysiology and drug
response. Precision medicine in SLE is trying to address
the need to assess SLE patients optimally, predict disease
course and treatment response at diagnosis. Ideally every
patient would undergo an initial evaluation that would
profile his/her disease, assessing the main pathophysiolo-
gic pathway through biomarkers, ther
---
REVIEW
T owards Precision Medicine in Systemic Lupus
Erythematosus…
=== kg_rels ===
Systemic lupus erythematosus - AUTHORED(Published in) -> N. Engl. J. Med.
---
Lisnevskaia L - AUTHORED() -> Systemic lupus erythematosus
---
Murphy G - AUTHORED() -> Systemic lupus erythematosus
---
Isenberg DA - AUTHORED() -> Systemic lupus erythematosus
---
Systemic lupus erythematosus - CITES(Published in) -> Lancet
---
Systemic lupus erythematosus - CITES(Systemic lupus erythematosus is discussed in the Lancet publication.) -> Lancet
---
Systemic lupus erythematosus - ASSOCIATED_WITH(SLE is characterized by aberrant activity of the immune system) -> Aberrant activity of the immune system
---
Immunological biomarkers - USED_FOR(Immunological biomarkers could diagnose and monitor disease activity in SLE) -> Systemic lupus erythematosus
---
Novel SLE biomarkers - DESCRIBES(Novel SLE biomarkers have been discovered through omics research) -> Systemic lupus erythematosus
—
…
Visualized in the console, we are returning the below subgraph, a combination of the text chunks from vector search, plus the entities and connected knowledge graph relationships.
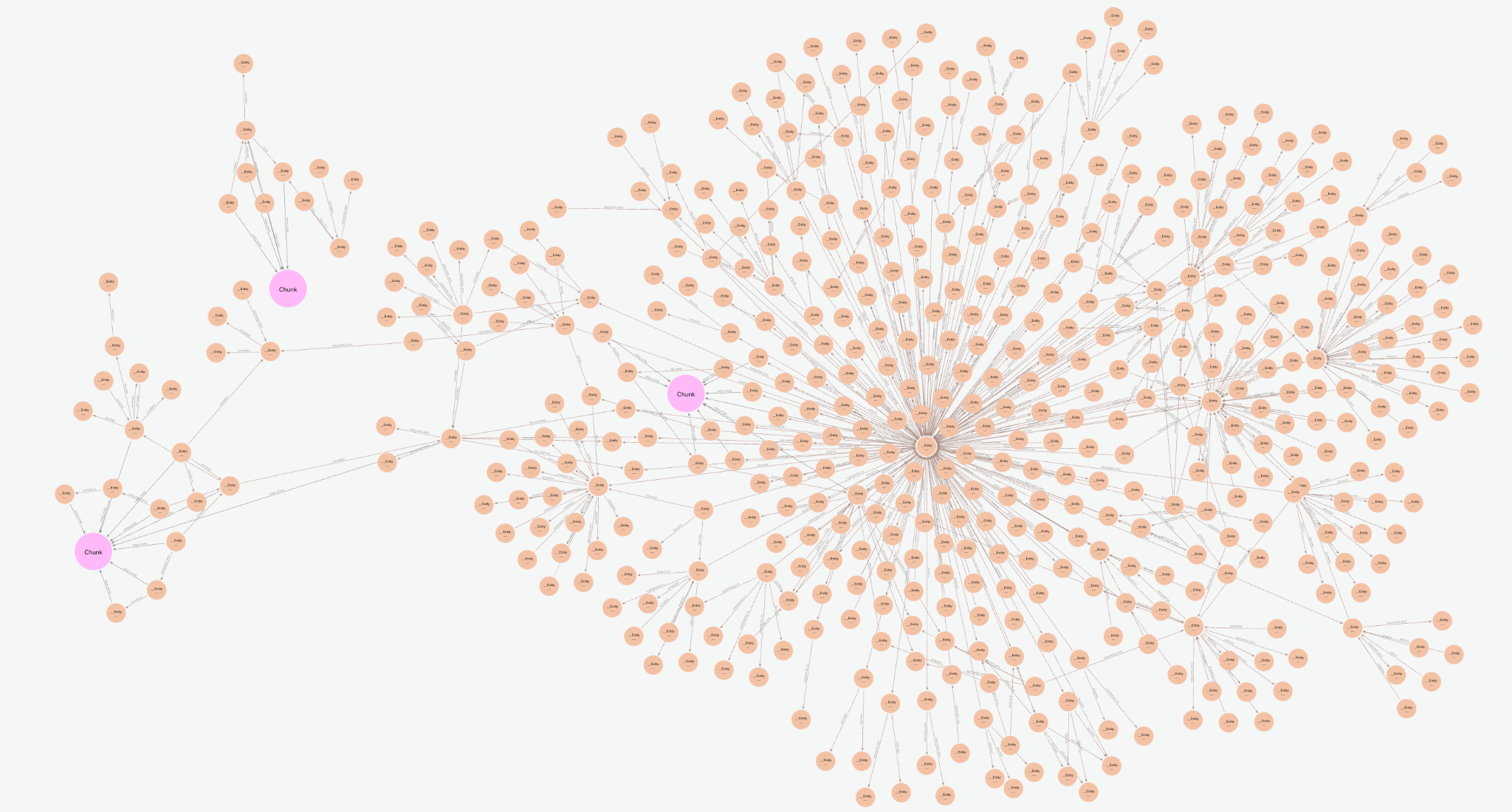
The vector cypher retriever is textualizing the entities graph by returning relationship sets in the format (node)-[rel]->(node). These connect multiple chunks and potentially multiple documents, combining insights from various parts of the database. Instead of just looking at individual text chunks, the relationships between entities can help reveal broader, interconnected information, enabling us to answer more complex queries later on. The explicit nature of the relationships also provides some enhanced explainability as one can see precisely how facts are derived. One can also iterate on the quality of these relationships to better fit their use case by tuning the KG construction covered above.
In the next section, we’ll create a GraphRAG object that we can invoke to see how the different retrievers behave.
3. Instantiate and Run GraphRAG
The GraphRAG Python package makes instantiating and running GraphRAG pipelines easy. We can use a dedicated GraphRAG class. At a minimum, you need to pass the constructor an LLM and a retriever. You can optionally pass a custom prompt template. We will do so here, just to provide a bit more guidance for the LLM to stick to information from our data source.
Below we create GraphRAG objects for both the vector and vector-cypher retrievers.
from neo4j_graphrag.llm import OpenAILLM as LLM
from neo4j_graphrag.generation import RagTemplate
from neo4j_graphrag.generation.graphrag import GraphRAG
llm = LLM(model_name="gpt-4o", model_params={"temperature": 0.0})
rag_template = RagTemplate(template='''Answer the Question using the following Context. Only respond with information mentioned in the Context. Do not inject any speculative information not mentioned.
# Question:
{query_text}
# Context:
{context}
# Answer:
''', expected_inputs=['query_text', 'context'])
v_rag = GraphRAG(llm=llm, retriever=vector_retriever, prompt_template=rag_template)
vc_rag = GraphRAG(llm=llm, retriever=vc_retriever, prompt_template=rag_template)
Now we can ask a simple question and see how the different knowledge graph retrieval patterns compare:
q = "How is precision medicine applied to Lupus? provide in list format."
print(f"Vector Response: n{v_rag.search(q, retriever_config={'top_k':5}).answer}")
print("n===========================n")
print(f"Vector + Cypher Response: n{vc_rag.search(q, retriever_config={'top_k':5}).answer}")
| Vector RAG Response | Vector + Cypher RAG Response |
|---|---|
|
|
Answers to simple questions like these can be similar between different knowledge graph vector retrieval approaches. We see that both answers contain similar information. For other simple questions, you may find Vector responses slightly more generic or broader, with Vector + Cypher responses being a bit more domain-specific or technical due to it sourcing the domain/entities portion of the knowledge graph, which was specifically structured for medical research (if you look back at our entity extraction prompt and schema).
Of course, one can tune and combine retrieval methods to further improve these responses; this is just a starting example. Let’s ask a bit more complex questions that require sourcing information from multiple text chunks.
q = "Can you summarize systemic lupus erythematosus (SLE)? including common effects, biomarkers, and treatments? Provide in detailed list format."
v_rag_result = v_rag.search(q, retriever_config={'top_k': 5}, return_context=True)
vc_rag_result = vc_rag.search(q, retriever_config={'top_k': 5}, return_context=True)
print(f"Vector Response: n{v_rag_result.answer}")
print("n===========================n")
print(f"Vector + Cypher Response: n{vc_rag_result.answer}")
Output:
| Vector RAG Response | Vector + Cypher RAG Response |
|---|---|
Systemic Lupus Erythematosus (SLE) Overview:
Common Effects:
Biomarkers:
Treatments:
|
Systemic Lupus Erythematosus (SLE) Summary:
Common Effects:
Biomarkers:
Treatments:
Management Strategies:
Note: |
These answers show a more significant difference. The Vector + Cypher response is more detailed and specific, particularly about biomarkers and treatments, making it more useful for an expert seeking depth on the subject. The Vector response is reasonably well structured and has a good overview of common effects, but it isn’t complete with technical details. It doesn’t contain a lot of domain-specific, relevant information from the documents.
In this case, the Vector + Cypher response uses entities and relationships in the knowledge graph that connect information from multiple documents and text chunks, making it more effective for questions that demand information from multiple sources.
Comparing the context returned, we can see that the vector-only retriever returned systematically similar text chunks, even containing the specific word “biomarker.” However, they do not contain certain technical details, like specific biomarkers.
for i in v_rag_result.retriever_result.items: print(json.dumps(eval(i.content), indent=1))
#output
{
"text": "ld be noted that nreproducibility and reliability may be affected by laboratory errors, specific techniques, nor changes in storage [13]. Because SLE can cause damage to various organs, has a complex pathogenesis, and displays heterogeneous clinical manifestations, one nparticular biomarker may only reflect one specific aspect of SLE but not be useful for nreflecting the state of the disease as a whole [14,15]. n nFigure 1. Common biomarkers for SLE and their measur ement sites in patients with "
}
{
"text": "agnosed and classified based on a patientu2019s clinical symptoms, signs, and nlaboratory biomarkers that reflect immune reactivity and inflammation in various norgans. It is necessary to develop consistent cl assification criteria of SLE for research and nclinical diagnosis. The most widely used classification criteria for SLE was established by the American College of Rheumatology (ACR) and contains laboratory biomarkers, nFigure 1. Common biomarkers for SLE and their measurement sites in patient"
}
{
"text": "s nElena Elefantea, Alain Cornetb, Jeanette Andersenb, Amy Somersb, Marta Moscaa,* naRheumatology Unit, Department of Clinical and Experimental Medicine, Azienda Ospedaliero-Universitaria Pisana, Italy nbLupus Europe, Brussels, Belgium nARTICLE INFO nKeywords: nSystemic Lupus Erythematosus nPatient-reported outcomes nPatient-clinician communication gap nHealth-related quality of life ABSTRACT nSystemic Lupus Erythematosus (SLE) imposes a great burden on the lives of patients. Patients u2019 and "
}
{
"text": "immunological biomarkers that could diagnose and monitor disease activity in SLE, with andnwithout organ-speciufb01c injury. In addition, novel SLE biomarkers that have been discovered throughnu201comicsu201d research are also reviewed.nKeywords: systemic lupus erythematosus; biomarkers; diagnosis; monitoring; omicsn1. IntroductionnSystemic lupus erythematosus (SLE) is a systemic autoimmune disease characterizednby aberrant activity of the immune system [ 1] and presents with a wide range of clinicalnmanife"
}
{
"text": "nSystemic Lupus Erythematosus (SLE) imposes a great burden on the lives of patients. Patients u2019 and nphysicians u2019 concerns about the disease diverge considerably. Physicians focus on controlling ndisease activity to prevent damage accrual, while patients focus on symptoms that impact on nHealth-Related Quality of Life (HRQoL). We explored the physicians u2019 and patients u2019 perspective nand the potential role of Patient Reported Outcomes (PROs). Physicians are aware of the theo-nretical usefulness o"
}
On the other hand, if we search for biomarkers in the context returned by Vector + Cypher we will see the relationships covering the domain-specific context. Below is a sample from the output:
vc_ls = vc_rag_result.retriever_result.items[0].content.split('\n---\n')
for i in vc_ls:
if "biomarker" in i: print(i)
#output (sample)
…
IP-10 - BIOMARKER_FOR(IP-10 is a biomarker for SLE) -> SLE
…
ANA - BIOMARKER_FOR(ANA is highly characteristic of SLE and can be used as a biomarker.) -> SLE
anti-Sm antibodies - BIOMARKER_FOR(Presence of anti-Sm antibodies serves as a biomarker for SLE classification) -> SLE
…
dysfunctional high-density lipoprotein - BIOMARKER_FOR(dysfunctional high-density lipoprotein is a key biomarker for SLE patients with CVD) -> SLE
Anti-dsDNA antibodies - ASSOCIATED_WITH(Anti-dsDNA antibodies are biomarkers associated with SLE.) -> SLE
…
You can repeat this for treatments and see the same pattern.
Ultimately, the GraphRAG Python package provides these and multiple other data retrieval methods you can tune, combine, and customize. Thus, you have comprehensive options for designing GraphRAG pipelines in your GenAI application.
GraphRAG for Beginners
Build a GraphRAG application that can answer complex questions based on connected data. Learn the three major retrieval patterns.
Resources to Simplify Your GraphRAG Experience
The GraphRAG Python package makes it easier than ever to go from documents to knowledge graph creation and retrieval—and you don’t need to be a Neo4j expert or write a lot of code. We’ve also compiled a series of guides and classes that feature common GraphRAG patterns and will help simplify your development experience. Be sure to check out these resources:
- GraphRAG Python package documentation
- In-depth blog posts:
- Getting Started With a Structured Knowledge Graph
- Enriching Vector Search With Graph Traversal (featuring VectorCypherRetriever)
- Hybrid Retrieval for GraphRAG Applications (hybrid vector/text search)
- Hybrid Vector/Text Search with Cypher Traversals
- Other learning resources for GraphRAG & GenAI – including Graph Academy and coded examples: